What's a DataFrame?¶
dataframe.head()
| hours_worked | wage_per_hour | |
|---|---|---|
| person_id | ||
| aafee0b | 38.5 | 15.1 |
| 0a917a3 | 41.25 | 15.0 |
| 2d1786b | 35.0 | 21.3 |
| 263cf89 | 27.75 | 17.5 |
| 89a09dc | 22.25 | 19.5 |
What's Data Validation?¶
Data validation is the act of falsifying data against explicit assumptions for some downstream purpose, like analysis, modeling, and visualization.
"All swans are white"
Why Do I Need it?¶
- It can be difficult to reason about and debug data processing pipelines.
- It's critical to ensuring data quality in many contexts especially when the end product informs business decisions, supports scientific findings, or generates predictions in a production setting.
Everyone has a personal relationship with their dataframes¶
One day, you encounter an error log trail and decide to follow it...¶
/usr/local/miniconda3/envs/pandera-presentations/lib/python3.7/site-packages/pandas/core/ops/__init__.py in masked_arith_op(x, y, op)
445 if mask.any():
446 with np.errstate(all="ignore"):
--> 447 result[mask] = op(xrav[mask], com.values_from_object(yrav[mask]))
448
449 else:
TypeError: can't multiply sequence by non-int of type 'float'
And you find yourself at the top of a function...¶
def process_data(df):
...
You look around, and see some hints of what had happened...¶
def process_data(df):
return (
df.assign(
weekly_income=lambda x: x.hours_worked * x.wage_per_hour
)
)
You sort of know what's going on, but you want to take a closer look!¶
def process_data(df):
import pdb; pdb.set_trace() # <- insert breakpoint
return (
df.assign(
weekly_income=lambda x: x.hours_worked * x.wage_per_hour
)
)
And you find some funny business going on...¶
>>> print(df)
hours_worked wage_per_hour person_id aafee0b 38.5 15.1 0a917a3 41.25 15.0 2d1786b 35.0 21.3 263cf89 27.75 17.5 89a09dc 22.25 19.5 e256747 -20.5 25.5
>>> df.dtypes
hours_worked object wage_per_hour float64 dtype: object
>>> df.hours_worked.map(type)
person_id aafee0b <class 'float'> 0a917a3 <class 'float'> 2d1786b <class 'str'> 263cf89 <class 'float'> 89a09dc <class 'float'> e256747 <class 'float'> Name: hours_worked, dtype: object
You squash the bug and add documentation for the next weary traveler who happens upon this code.¶
def process_data(df):
return (
df
.astype({"hours_worked": float, "wage_per_hour": float}) # <- make sure columns are floats
.assign(
hours_worked=lambda x: x.hours_worked.where( # <- replace negative values with nans
x.hours_worked >= 0, np.nan
)
)
.assign(
weekly_income=lambda x: x.hours_worked * x.wage_per_hour
)
)
>>> process_data(df)
| hours_worked | wage_per_hour | weekly_income | |
|---|---|---|---|
| person_id | |||
| aafee0b | 38.50 | 15.1 | 581.350 |
| 0a917a3 | 41.25 | 15.0 | 618.750 |
| 2d1786b | 35.00 | 21.3 | 745.500 |
| 263cf89 | 27.75 | 17.5 | 485.625 |
| 89a09dc | 22.25 | 19.5 | 433.875 |
| e256747 | NaN | 25.5 | NaN |
A few months later...¶
You find yourself at a familiar function, but it looks a little different from when you left it...¶
@pa.check_input(in_schema)
@pa.check_output(out_schema)
def process_data(df):
return (
df.assign(
hours_worked=lambda x: x.hours_worked.where( # <- replace negative values with nans
x.hours_worked >= 0, np.nan
)
)
.assign(
weekly_income=lambda x: x.hours_worked * x.wage_per_hour
)
)
You look above and see what in_schema and out_schema are, finding a NOTE that a fellow traveler has left for you.¶
import pandera as pa
from pandera import DataFrameSchema, Column, Check
# NOTE: this is what's supposed to be in `df` going into `process_data`
in_schema = DataFrameSchema({
"hours_worked": Column(pa.Float, coerce=True, nullable=True),
"wage_per_hour": Column(pa.Float, coerce=True, nullable=True),
})
# ... and this is what `process_data` is supposed to return.
out_schema = (
in_schema
.update_column("hours_worked", checks=Check.greater_than_or_equal_to(0))
.add_columns({"weekly_income": Column(pa.Float, nullable=True)})
)
@pa.check_input(in_schema)
@pa.check_output(out_schema)
def process_data(df):
...
Moral of the Story¶
The better you can reason about the contents of a dataframe, the faster you can debug.
The faster you can debug, the sooner you can focus on downstream tasks that you care about.
Outline¶
- Data validation in theory and practice.
- Brief introduction to
pandera - Case Study: Fatal Encounters dataset
- Roadmap
Data Validation in Theory and Practice¶
According to the European Statistical System:
Data validation is an activity in which it is verified whether or not a combination of values is a member of a set of acceptable value combinations. [Di Zio et al. 2015]
Data Validation in Theory and Practice¶
More formally, we can relate this definition to one of the core principles of the scientific method: falsifiability
$v(x) \twoheadrightarrow \{ {True}, {False} \}$
Where $x$ is a set of data values and $v$ is a surjective validation function, meaning that there exists at least one $x$ that maps onto each of the elements in the set $\{True, False\}$ [van der Loo et al. 2019].
Case 1: Unfalsifiable¶
$v(x) \rightarrow True$
Example: "my dataframe can have any number of columns of any type"
lambda df: True
Case 2: Unverifiable¶
$v(x) \rightarrow False$
Example: "my dataframe has an infinite number of rows and columns"
lambda df: False
Types of Validation Rules¶
Technical Checks¶
Have to do with the properties of the data structure:
incomeis a numeric variable,occupationis a categorical variable.email_addressshould be unique.- null (missing) values are permitted in the
occupationfield.
Domain-specific Checks¶
Have to do with properties specific to the topic under study:
- the
agevariable should be between the range 0 and 120 - the
incomefor records whereageis below the legal working age should benan - certain categories of
occupationtend to have higherincomethan others
Types of Validation Rules¶
Deterministic Checks¶
Checks that express hard-coded logical rules
the mean
ageshould be between30and40years old.lambda age: 30 <= age.mean() <= 40
Probabilistic Checks¶
Checks that explicitly incorporate randomness and distributional variability
the 95% confidence interval or mean
ageshould be between30and40years old.def prob_check_age(age): mu = age.mean() ci = 1.96 * (age.std() / np.sqrt(len(age)) return 30 <= mu - ci and mu + ci <= 40
Statistical Type Safety¶
Verifying the assumptions about the distributional properties of a dataset to ensure that statistical operations on those data are valid.
- "the training samples are independently and identically distributed"
- "features
x1,x2, andx3are not correlated"
- "the variance of feature
xis greater than some thresholdt"
- "the label distribution is consistent across the training and test set"
Data Validation Workflow in Practice¶
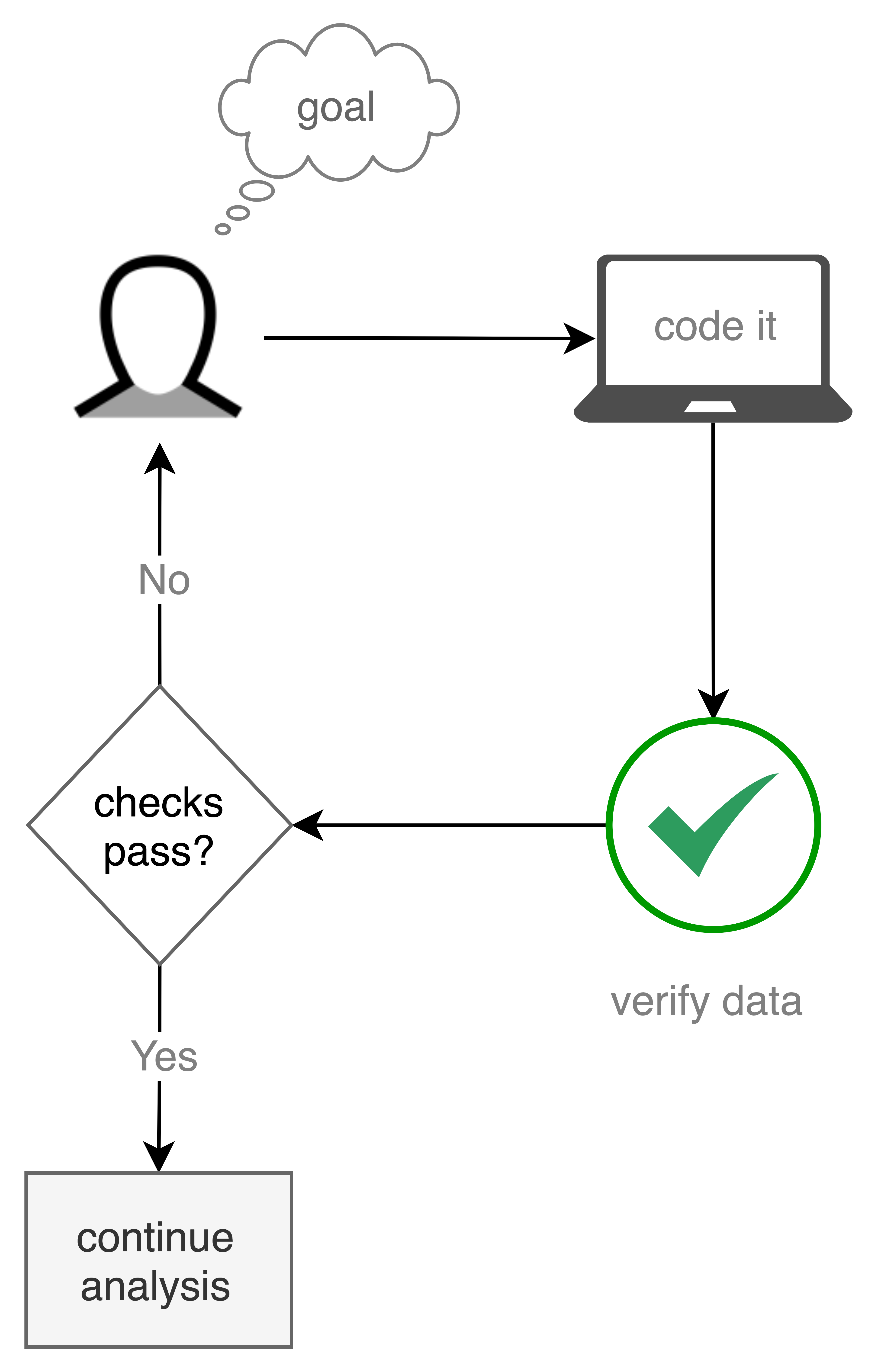
User Story¶
As a machine learning engineer who uses
pandasevery day, I want a data validation tool that's intuitive, flexible, customizable, and easy to integrate into my ETL pipelines so that I can spend less time worrying about the correctness of a dataframe's contents and more time training models.

Introducing pandera¶
A design-by-contract data validation library that exposes an intuitive API for expressing dataframe schemas.
Refactoring the Validation Function¶
$v(x) \twoheadrightarrow \{ {True}, {False} \}$
$s(v, x) \rightarrow \begin{cases} \mbox{x,} & \mbox{if } v(x) = true \\ \mbox{error,} & \mbox{otherwise} \end{cases}$
Where $s$ is a schema function that takes two arguments: the validation function $v$ and some data $x$.
Why?¶
Compositionality
Consider a data processing function $f(x) \rightarrow x'$ that cleans the raw dataset $x$.
We can use the schema to define any number of composite functions:
- $f(s(x))$ first validates the raw data to catch invalid data before it's preprocessed.
- $s(f(x))$ validates the output of $f$ to check that the processing function is fulfilling the contract defined in $s$
- $s(f(s'(x))$: the data validation 🥪
Architecture¶
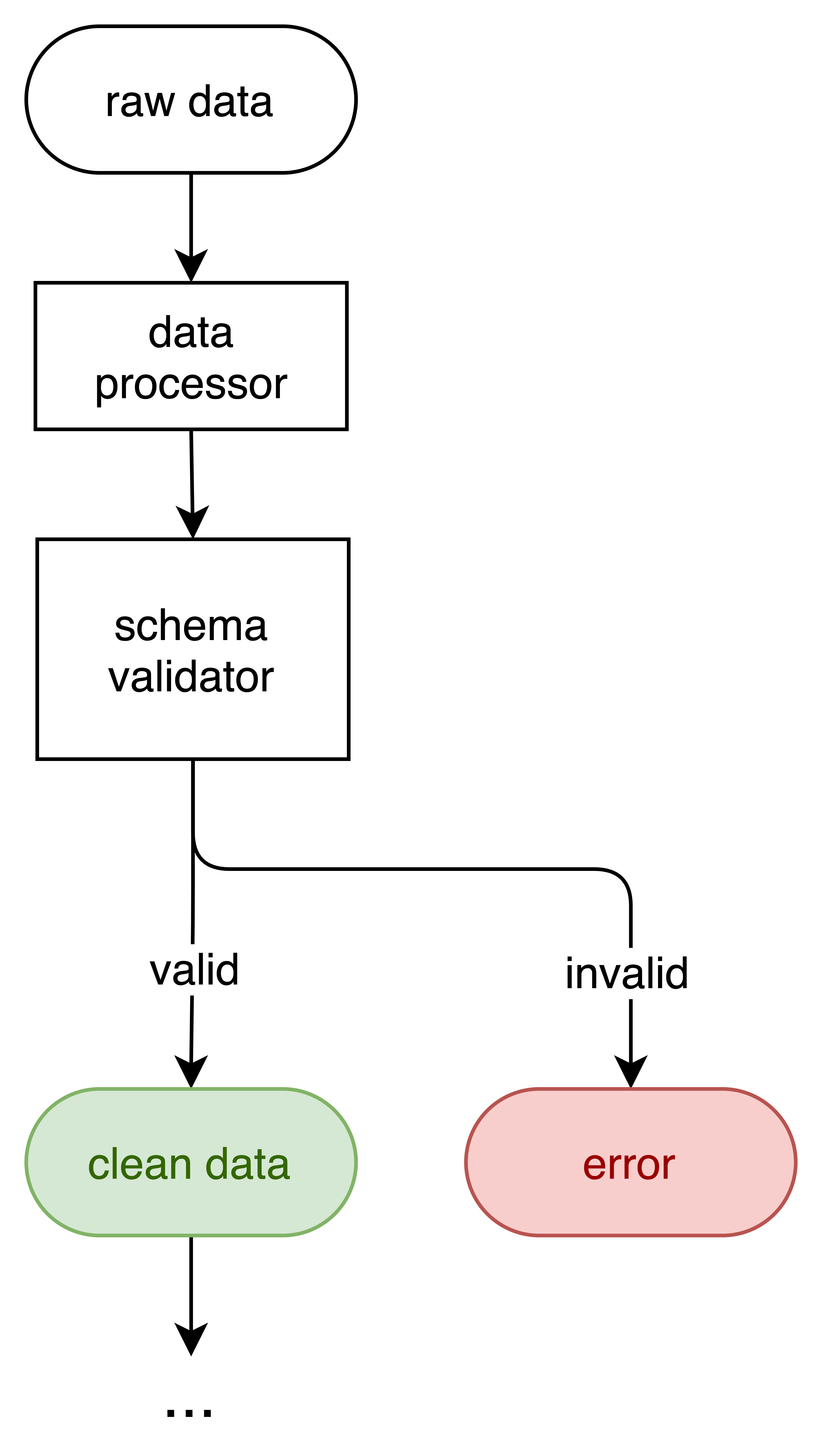
Case Study: Fatal Encounters Dataset¶
"A step toward creating an impartial, comprehensive, and searchable national database of people killed during interactions with law enforcement."
Bird's Eye View¶
- 26,000+ records of law enforcement encounters leading to death
- Records date back to the year 2000
- Each record contains:
- demographics of the decedent
- cause and location of the death
- agency responsible for death
- court disposition of the case (e.g. "Justified", "Accidental", "Suicide", "Criminal")
What factors are most predictive of the court ruling a case as "Accidental"?¶
Example 1:
Undocumented immigrant Roberto Chavez-Recendiz, of Hidalgo, Mexico, was fatally shot while Rivera was arresting him along with his brother and brother-in-law for allegedly being in the United States without proper documentation. The shooting was "possibly the result of what is called a 'sympathetic grip,' where one hand reacts to the force being used by the other," Chief Criminal Deputy County Attorney Rick Unklesbay wrote in a letter outlining his review of the shooting. The Border Patrol officer had his pistol in his hand as he took the suspects into custody. He claimed the gun fired accidentally.
Example 2:
Andrew Lamar Washington died after officers tasered him 17 times within three minutes.
Example 3:
Biddle and his brother, Drake Biddle, were fleeing from a Nashville Police Department officer at a high rate of speed when a second Nashville Police Department officer, James L. Steely, crashed into him head-on.
Caveats¶
- As mentioned on the website, the records in this dataset are the most comprehensive to date, but by no means is it a finished project. Biases may be lurking everywhere!
- I don't have any domain expertise in criminal justice!
- The main purpose here is to showcase the capabilities of
pandera.

Notebook is Available Here:¶
Read the Data¶
import janitor
import requests
from pandas_profiling import ProfileReport
dataset_url = (
"https://docs.google.com/spreadsheets/d/"
"1dKmaV_JiWcG8XBoRgP8b4e9Eopkpgt7FL7nyspvzAsE/export?format=csv"
)
fatal_encounters = pd.read_csv(dataset_url, skipfooter=1, engine="python")
| Unique ID | Subject's name | Subject's age | Subject's gender | Subject's race | Subject's race with imputations | Imputation probability | URL of image of deceased | Date of injury resulting in death (month/day/year) | Location of injury (address) | Location of death (city) | Location of death (state) | Location of death (zip code) | Location of death (county) | Full Address | Latitude | Longitude | Agency responsible for death | Cause of death | A brief description of the circumstances surrounding the death | Dispositions/Exclusions INTERNAL USE, NOT FOR ANALYSIS | Intentional Use of Force (Developing) | Link to news article or photo of official document | Symptoms of mental illness? INTERNAL USE, NOT FOR ANALYSIS | Video | Date&Description | Unique ID formula | Unique identifier (redundant) | Date (Year) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 25746 | Samuel H. Knapp | 17 | Male | European-American/White | European-American/White | not imputed | NaN | 01/01/2000 | 27898-27804 US-101 | Willits | CA | 95490.0 | Mendocino | 27898-27804 US-101 Willits CA 95490 Mendocino | 39.470883 | -123.361751 | Mendocino County Sheriff's Office | Vehicle | Samuel Knapp was allegedly driving a stolen ve... | Unreported | Vehicle/Pursuit | https://drive.google.com/file/d/10DisrV8K5ReP1... | No | NaN | 1/1/2000: Samuel Knapp was allegedly driving a... | NaN | 25746 | 2000.0 |
| 1 | 25747 | Mark A. Horton | 21 | Male | African-American/Black | African-American/Black | not imputed | NaN | 01/01/2000 | Davison Freeway | Detroit | MI | 48203.0 | Wayne | Davison Freeway Detroit MI 48203 Wayne | 42.404526 | -83.092274 | NaN | Vehicle | Two Detroit men killed when their car crashed ... | Unreported | Vehicle/Pursuit | https://drive.google.com/file/d/1-nK-RohgiM-tZ... | No | NaN | 1/1/2000: Two Detroit men killed when their ca... | NaN | 25747 | 2000.0 |
| 2 | 25748 | Phillip A. Blurbridge | 19 | Male | African-American/Black | African-American/Black | not imputed | NaN | 01/01/2000 | Davison Freeway | Detroit | MI | 48203.0 | Wayne | Davison Freeway Detroit MI 48203 Wayne | 42.404526 | -83.092274 | NaN | Vehicle | Two Detroit men killed when their car crashed ... | Unreported | Vehicle/Pursuit | https://drive.google.com/file/d/1-nK-RohgiM-tZ... | No | NaN | 1/1/2000: Two Detroit men killed when their ca... | NaN | 25748 | 2000.0 |
Clean up column names¶
To make the analysis more readable
def clean_columns(df):
return (
df.clean_names()
.rename(
columns=lambda x: (
x.strip("_")
.replace("&", "_and_")
.replace("subjects_", "")
.replace("location_of_death_", "")
.replace("_resulting_in_death_month_day_year", "")
.replace("_internal_use_not_for_analysis", "")
)
)
)
fatal_encounters_clean_columns = clean_columns(fatal_encounters)
Minimal Schema Definition¶
Just define columns that we need for creating the training set,
and specify which are nullable.
clean_column_schema = pa.DataFrameSchema(
{
"age": Column(nullable=True),
"gender": Column(nullable=True),
"race": Column(nullable=True),
"cause_of_death": Column(nullable=True),
"symptoms_of_mental_illness": Column(nullable=True),
"dispositions_exclusions": Column(nullable=True),
}
)
Validate the output of clean_columns using pipe at the end of the method chain
def clean_columns(df):
return (
df.clean_names()
.rename(
columns=lambda x: (
x.strip("_")
.replace("&", "_and_")
.replace("subjects_", "")
.replace("location_of_death_", "")
.replace("_resulting_in_death_month_day_year", "")
.replace("_internal_use_not_for_analysis", "")
)
)
.pipe(clean_column_schema) # <- validate output with schema
)
Sidebar¶
If a column is not present as specified by the schema, a SchemaError is raised.
corrupted_data = fatal_encounters.drop("Subject's age", axis="columns")
try:
clean_columns(corrupted_data)
except pa.errors.SchemaError as exc:
print(exc)
column 'age' not in dataframe
unique_id name gender race \
0 25746 Samuel H. Knapp Male European-American/White
1 25747 Mark A. Horton Male African-American/Black
2 25748 Phillip A. Blurbridge Male African-American/Black
3 25749 Mark Ortiz Male Hispanic/Latino
4 2 Lester Miller Male Race unspecified
race_with_imputations imputation_probability url_of_image_of_deceased \
0 European-American/White not imputed NaN
1 African-American/Black not imputed NaN
2 African-American/Black not imputed NaN
3 Hispanic/Latino not imputed NaN
4 African-American/Black 0.947676492 NaN
date_of_injury location_of_injury_address city ... \
0 01/01/2000 27898-27804 US-101 Willits ...
1 01/01/2000 Davison Freeway Detroit ...
2 01/01/2000 Davison Freeway Detroit ...
3 01/01/2000 600 W Cherry Ln Carlsbad ...
4 01/02/2000 4850 Flakes Mill Road Ellenwood ...
a_brief_description_of_the_circumstances_surrounding_the_death \
0 Samuel Knapp was allegedly driving a stolen ve...
1 Two Detroit men killed when their car crashed ...
2 Two Detroit men killed when their car crashed ...
3 A motorcycle was allegedly being driven errati...
4 Darren Mayfield, a DeKalb County sheriff's dep...
dispositions_exclusions intentional_use_of_force_developing \
0 Unreported Vehicle/Pursuit
1 Unreported Vehicle/Pursuit
2 Unreported Vehicle/Pursuit
3 Unreported Vehicle/Pursuit
4 Criminal Intentional Use of Force, Deadly
link_to_news_article_or_photo_of_official_document \
0 https://drive.google.com/file/d/10DisrV8K5ReP1...
1 https://drive.google.com/file/d/1-nK-RohgiM-tZ...
2 https://drive.google.com/file/d/1-nK-RohgiM-tZ...
3 https://drive.google.com/file/d/1qAEefRjX_aTtC...
4 https://docs.google.com/document/d/1-YuShSarW_...
symptoms_of_mental_illness video \
0 No NaN
1 No NaN
2 No NaN
3 No NaN
4 No NaN
date_and_description unique_id_formula \
0 1/1/2000: Samuel Knapp was allegedly driving a... NaN
1 1/1/2000: Two Detroit men killed when their ca... NaN
2 1/1/2000: Two Detroit men killed when their ca... NaN
3 1/1/2000: A motorcycle was allegedly being dri... NaN
4 1/2/2000: Darren Mayfield, a DeKalb County she... NaN
unique_identifier_redundant date_year
0 25746 2000.0
1 25747 2000.0
2 25748 2000.0
3 25749 2000.0
4 2 2000.0
[5 rows x 28 columns]
Explore the Data with pandas-profiling¶
Declare the Training Data Schema¶
training_data_schema = pa.DataFrameSchema(
{
# feature columns
"age": Column(pa.Float, Check.in_range(0, 120), nullable=True),
"gender": Column(pa.String, Check.isin(genders), nullable=True),
"race": Column(pa.String, Check.isin(races), nullable=True),
"cause_of_death": Column(pa.String, Check.isin(causes_of_death), nullable=True),
"symptoms_of_mental_illness": Column(pa.Bool, nullable=True),
# target column
"disposition_accidental": Column(pa.Bool, nullable=False),
},
coerce=True # <- coerce columns to the specified type
)
Serialize schema to yaml format:¶
print(training_data_schema.to_yaml())
schema_type: dataframe
version: 0.4.2
columns:
age:
pandas_dtype: float
nullable: true
checks:
in_range:
min_value: 0
max_value: 120
gender:
pandas_dtype: string
nullable: true
checks:
isin:
- female
- male
- transgender
- transexual
race:
pandas_dtype: string
nullable: true
checks:
isin:
- african_american_black
- asian_pacific_islander
- european_american_white
- hispanic_latino
- middle_eastern
- native_american_alaskan
- race_unspecified
cause_of_death:
pandas_dtype: string
nullable: true
checks:
isin:
- asphyxiated_restrained
- beaten_bludgeoned_with_instrument
- burned_smoke_inhalation
- chemical_agent_pepper_spray
- drowned
- drug_overdose
- fell_from_a_height
- gunshot
- medical_emergency
- other
- stabbed
- tasered
- undetermined
- unknown
- vehicle
symptoms_of_mental_illness:
pandas_dtype: bool
nullable: true
checks: null
disposition_accidental:
pandas_dtype: bool
nullable: false
checks: null
index: null
coerce: true
Clean the Data¶
The cleaning function should normalize string values as specified
by training_data_schema.
def clean_data(df):
return (
df.dropna(subset=["dispositions_exclusions"])
.transform_columns(
[
"gender", "race", "cause_of_death",
"symptoms_of_mental_illness", "dispositions_exclusions"
],
lambda x: x.str.lower().str.replace('-|/| ', '_'), # clean string values
elementwise=False
)
.transform_column(
"symptoms_of_mental_illness",
lambda x: x.mask(x.dropna().str.contains("unknown")) != "no", # binarize mental illness
elementwise=False
)
.transform_column(
"dispositions_exclusions",
lambda x: x.str.contains("accident", case=False), # derive target column
"disposition_accidental",
elementwise=False
)
.query("gender != 'white'") # probably a data entry error
.filter_string(
"dispositions_exclusions",
"unreported|unknown|pending|suicide", # filter out unknown, unreported, or suicide cases
complement=True
)
)
Add the data validation 🥪¶
@pa.check_input(clean_column_schema)
@pa.check_output(training_data_schema)
def clean_data(df):
return (
df.dropna(subset=["dispositions_exclusions"])
.transform_columns(
[
"gender", "race", "cause_of_death",
"symptoms_of_mental_illness", "dispositions_exclusions"
],
lambda x: x.str.lower().str.replace('-|/| ', '_'), # clean string values
elementwise=False
)
.transform_column(
"symptoms_of_mental_illness",
lambda x: x.mask(x.dropna().str.contains("unknown")) != "no", # binarize mental illness
elementwise=False
)
.transform_column(
"dispositions_exclusions",
lambda x: x.str.contains("accident", case=False), # derive target column
"disposition_accidental",
elementwise=False
)
.query("gender != 'white'") # probably a data entry error
.filter_string(
"dispositions_exclusions",
"unreported|unknown|pending|suicide", # filter out unknown, unreported, or suicide cases
complement=True
)
)
ValueError: Unable to coerce column to schema data type¶
try:
clean_data(fatal_encounters_clean_columns)
except ValueError as exc:
print(exc)
could not convert string to float: '18 months'
Normalize the age column¶
So that string values can be converted into float
def normalize_age(age):
return (
age.str.replace("s|`", "")
.pipe(normalize_age_range)
.pipe(normalize_age_to_year, "month|mon", 12)
.pipe(normalize_age_to_year, "day", 365)
)
Apply normalize_age inside the clean_data function.¶
# data validation 🥪
@pa.check_input(clean_column_schema)
@pa.check_output(training_data_schema)
def clean_data(df):
return (
df.dropna(subset=["dispositions_exclusions"])
.transform_columns(
[
"gender", "race", "cause_of_death",
"symptoms_of_mental_illness", "dispositions_exclusions"
],
lambda x: x.str.lower().str.replace('-|/| ', '_'), # clean string values
elementwise=False
)
.transform_column(
"symptoms_of_mental_illness",
lambda x: x.mask(x.dropna().str.contains("unknown")) != "no", # binarize mental illness
elementwise=False
)
.transform_column("age", normalize_age, elementwise=False) # <- clean up age column
.transform_column(
"dispositions_exclusions",
lambda x: x.str.contains("accident", case=False), # derive target column
"disposition_accidental",
elementwise=False
)
.query("gender != 'white'") # probably a data entry error
.filter_string(
"dispositions_exclusions",
"unreported|unknown|pending|suicide", # filter out unknown, unreported, or suicide cases
complement=True
)
)
Create Training Set¶
fatal_encounters_clean = clean_data(fatal_encounters_clean_columns)
with pd.option_context("display.max_rows", 5):
display(fatal_encounters_clean.filter(list(training_data_schema.columns)))
| age | gender | race | cause_of_death | symptoms_of_mental_illness | disposition_accidental | |
|---|---|---|---|---|---|---|
| 4 | 53.0 | male | race_unspecified | gunshot | False | False |
| 8 | 42.0 | female | race_unspecified | vehicle | False | False |
| ... | ... | ... | ... | ... | ... | ... |
| 28236 | 35.0 | male | european_american_white | drowned | False | False |
| 28276 | 26.0 | male | hispanic_latino | gunshot | False | False |
8068 rows × 6 columns
corrupt_data = fatal_encounters_clean.copy()
corrupt_data["gender"].iloc[:50] = "foo"
corrupt_data["gender"].iloc[50:100] = "bar"
try:
training_data_schema(corrupt_data)
except pa.errors.SchemaError as exc:
print(exc)
<Schema Column: 'gender' type=string> failed element-wise validator 0:
<Check _isin: isin({'transgender', 'female', 'male', 'transexual'})>
failure cases:
index count
failure_case
bar [146, 147, 148, 151, 153, 162, 165, 168, 171, ... 50
foo [4, 8, 9, 11, 12, 13, 17, 22, 31, 32, 40, 41, ... 50
Fine-grained debugging¶
The SchemaError exception object contains the invalid dataframe and
the failure cases, which is also a dataframe.
with pd.option_context("display.max_rows", 5):
try:
training_data_schema(corrupt_data)
except pa.errors.SchemaError as exc:
print("Invalid Data:\n-------------")
print(exc.data.iloc[:, :5])
print("\nFailure Cases:\n--------------")
print(exc.failure_cases)
Invalid Data:
-------------
unique_id name age gender \
4 2 Lester Miller 53.0 foo
8 25752 Doris Murphy 42.0 foo
... ... ... ... ...
28236 28209 Joe Deewayne Cothrum 35.0 male
28276 28358 Jose Santos Parra Juarez 26.0 male
race
4 race_unspecified
8 race_unspecified
... ...
28236 european_american_white
28276 hispanic_latino
[8068 rows x 5 columns]
Failure Cases:
--------------
index failure_case
0 4 foo
1 8 foo
.. ... ...
98 280 bar
99 284 bar
[100 rows x 2 columns]
Summarize the Data¶
What percent of cases in the training data are "accidental"?
percent_accidental = fatal_encounters_clean.disposition_accidental.mean()
display(Markdown(f"{percent_accidental * 100:0.02f}%"))
2.74%
Hypothesis: "the disposition_accidental target has a
class balance of ~2.75%"
from pandera import Hypothesis
# use the Column object as a stand-alone schema object
target_schema = Column(
pa.Bool,
name="disposition_accidental",
checks=Hypothesis.one_sample_ttest(
popmean=0.0275, relationship="equal", alpha=0.01
)
)
target_schema(fatal_encounters_clean);
Prepare Training and Test Sets¶
For functions that have tuple/list-like output, specify an integer
index pa.check_output(schema, <int>) to apply the schema to a
specific element in the output.
from sklearn.model_selection import train_test_split
target_schema = pa.SeriesSchema(
pa.Bool,
name="disposition_accidental",
checks=Hypothesis.one_sample_ttest(
popmean=0.0275, relationship="equal", alpha=0.01
)
)
feature_schema = training_data_schema.remove_columns([target_schema.name])
@pa.check_input(training_data_schema)
@pa.check_output(feature_schema, 0)
@pa.check_output(feature_schema, 1)
@pa.check_output(target_schema, 2)
@pa.check_output(target_schema, 3)
def split_training_data(fatal_encounters_clean):
return train_test_split(
fatal_encounters_clean[list(feature_schema.columns)],
fatal_encounters_clean[target_schema.name],
test_size=0.2,
random_state=45,
)
X_train, X_test, y_train, y_test = split_training_data(fatal_encounters_clean)
Model the Data¶
Import the tools
from sklearn.calibration import CalibratedClassifierCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.impute import SimpleImputer
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.compose import ColumnTransformer
DataFrameSchema -> ColumnTransformer 🤯¶
Create a transformer to numericalize the features using a schema object.
Checks have a statistics attribute that enables access to the properties
defined in the schema.
def column_transformer_from_schema(feature_schema):
def transformer_from_column(column):
column_schema = feature_schema.columns[column]
if column_schema.pandas_dtype is pa.String:
return make_pipeline(
SimpleImputer(strategy="most_frequent"),
OneHotEncoder(categories=[get_categories(column_schema)])
)
if column_schema.pandas_dtype is pa.Bool:
return SimpleImputer(strategy="median")
# otherwise assume numeric variable
return make_pipeline(
SimpleImputer(strategy="median"),
StandardScaler()
)
return ColumnTransformer([
(column, transformer_from_column(column), [column])
for column in feature_schema.columns
])
def get_categories(column_schema):
for check in column_schema.checks:
if check.name == "isin":
return check.statistics["allowed_values"]
raise ValueError("could not find Check.isin")
Define the transformer¶
transformer = column_transformer_from_schema(feature_schema)
ColumnTransformer(transformers=[('age',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='median')),
('standardscaler',
StandardScaler())]),
['age']),
('gender',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(categories=[['female',
'male',
'transgender',
'transexual']]))]),
['gender']),
('race',...
'beaten_bludgeoned_with_instrument',
'burned_smoke_inhalation',
'chemical_agent_pepper_spray',
'drowned',
'drug_overdose',
'fell_from_a_height',
'gunshot',
'medical_emergency',
'other',
'stabbed',
'tasered',
'undetermined',
'unknown',
'vehicle']]))]),
['cause_of_death']),
('symptoms_of_mental_illness',
SimpleImputer(strategy='median'),
['symptoms_of_mental_illness'])])['age']
SimpleImputer(strategy='median')
StandardScaler()
['gender']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['female', 'male', 'transgender', 'transexual']])
['race']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['african_american_black', 'asian_pacific_islander',
'european_american_white', 'hispanic_latino',
'middle_eastern', 'native_american_alaskan',
'race_unspecified']])['cause_of_death']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['asphyxiated_restrained',
'beaten_bludgeoned_with_instrument',
'burned_smoke_inhalation',
'chemical_agent_pepper_spray', 'drowned',
'drug_overdose', 'fell_from_a_height', 'gunshot',
'medical_emergency', 'other', 'stabbed', 'tasered',
'undetermined', 'unknown', 'vehicle']])['symptoms_of_mental_illness']
SimpleImputer(strategy='median')
Define and fit the modeling pipeline¶
You can even decorate object methods, specifying the argument name that you want to apply a schema to.
pipeline = Pipeline([
("transformer", transformer),
(
"estimator",
RandomForestClassifier(
class_weight="balanced_subsample",
n_estimators=500,
min_samples_leaf=20,
min_samples_split=10,
max_depth=10,
random_state=100,
)
)
])
fit_fn = pa.check_input(feature_schema, "X")(pipeline.fit)
fit_fn = pa.check_input(target_schema, "y")(fit_fn)
fit_fn(X_train, y_train)
Pipeline(steps=[('transformer',
ColumnTransformer(transformers=[('age',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='median')),
('standardscaler',
StandardScaler())]),
['age']),
('gender',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(categories=[['female',
'male',
'transgender',
'transex...
'medical_emergency',
'other',
'stabbed',
'tasered',
'undetermined',
'unknown',
'vehicle']]))]),
['cause_of_death']),
('symptoms_of_mental_illness',
SimpleImputer(strategy='median'),
['symptoms_of_mental_illness'])])),
('estimator',
RandomForestClassifier(class_weight='balanced_subsample',
max_depth=10, min_samples_leaf=20,
min_samples_split=10, n_estimators=500,
random_state=100))])ColumnTransformer(transformers=[('age',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='median')),
('standardscaler',
StandardScaler())]),
['age']),
('gender',
Pipeline(steps=[('simpleimputer',
SimpleImputer(strategy='most_frequent')),
('onehotencoder',
OneHotEncoder(categories=[['female',
'male',
'transgender',
'transexual']]))]),
['gender']),
('race',...
'beaten_bludgeoned_with_instrument',
'burned_smoke_inhalation',
'chemical_agent_pepper_spray',
'drowned',
'drug_overdose',
'fell_from_a_height',
'gunshot',
'medical_emergency',
'other',
'stabbed',
'tasered',
'undetermined',
'unknown',
'vehicle']]))]),
['cause_of_death']),
('symptoms_of_mental_illness',
SimpleImputer(strategy='median'),
['symptoms_of_mental_illness'])])['age']
SimpleImputer(strategy='median')
StandardScaler()
['gender']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['female', 'male', 'transgender', 'transexual']])
['race']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['african_american_black', 'asian_pacific_islander',
'european_american_white', 'hispanic_latino',
'middle_eastern', 'native_american_alaskan',
'race_unspecified']])['cause_of_death']
SimpleImputer(strategy='most_frequent')
OneHotEncoder(categories=[['asphyxiated_restrained',
'beaten_bludgeoned_with_instrument',
'burned_smoke_inhalation',
'chemical_agent_pepper_spray', 'drowned',
'drug_overdose', 'fell_from_a_height', 'gunshot',
'medical_emergency', 'other', 'stabbed', 'tasered',
'undetermined', 'unknown', 'vehicle']])['symptoms_of_mental_illness']
SimpleImputer(strategy='median')
RandomForestClassifier(class_weight='balanced_subsample', max_depth=10,
min_samples_leaf=20, min_samples_split=10,
n_estimators=500, random_state=100)Evaluate the Model¶
Use the check_input and check_output decorators to validate
the estimator.predict_proba method.
from sklearn.metrics import roc_auc_score, roc_curve
pred_schema = pa.SeriesSchema(pa.Float, Check.in_range(0, 1))
# check that the feature array input to predict_proba method adheres to the feature_schema
predict_fn = pa.check_input(feature_schema)(pipeline.predict_proba)
# check that the prediction array output is a probability.
predict_fn = pa.check_output(pred_schema, lambda x: pd.Series(x))(predict_fn)
yhat_train = pipeline.predict_proba(X_train)[:, 1]
print(f"train ROC AUC: {roc_auc_score(y_train, yhat_train):0.04f}")
yhat_test = pipeline.predict_proba(X_test)[:, 1]
print(f"test ROC AUC: {roc_auc_score(y_test, yhat_test):0.04f}")
train ROC AUC: 0.9207 test ROC AUC: 0.8479
Plot the ROC curves using an in-line schema.
def plot_roc_auc(y_true, y_pred, label, ax=None):
fpr, tpr, _ = roc_curve(y_true, y_pred)
roc_curve_df = pd.DataFrame({"fpr": fpr, "tpr": tpr}).pipe(
pa.DataFrameSchema({
"fpr": Column(pa.Float, Check.in_range(0, 1)),
"tpr": Column(pa.Float, Check.in_range(0, 1)),
})
)
return roc_curve_df.plot.line(x="fpr", y="tpr", label=label, ax=ax)
with sns.axes_style("whitegrid"):
_, ax = plt.subplots(figsize=(5, 4))
plot_roc_auc(y_train, yhat_train, "test AUC", ax)
plot_roc_auc(y_test, yhat_test, "train AUC", ax)
ax.set_ylabel("true_positive_rate")
ax.set_xlabel("false_positive_rate")
ax.plot([0, 1], [0, 1], color="k", linestyle=":")
Audit the Model¶
shap package: https://github.com/slundberg/shap
Create an explainer object. Here we want to check the inputs to the transformer.transform method.
import shap
explainer = shap.TreeExplainer(
pipeline.named_steps["estimator"],
feature_perturbation="tree_path_dependent",
)
transform_fn = pa.check_input(feature_schema)(
pipeline.named_steps["transformer"].transform
)
X_test_array = transform_fn(X_test).toarray()
shap_values = explainer.shap_values(X_test_array, check_additivity=False)
What factors are most predictive of the court ruling a case as "Accidental"?¶
The probability of the case being ruled as accidental ⬆️ if the cause_of_death is vehicle, tasered, asphyxiated_restrained, medical_emergency, or drug_overdose, or race is race_unspecified or native_american_alaskan.
The probability of the case being ruled as accidental ⬇️ if the cause_of_death is gunshot or race is european_american_white, or asian_pacific_islander.
Write the Model Audit Schema¶
Create a dataframe with {variable} and {variable}_shap as columns
audit_dataframe = (
pd.concat(
[
pd.DataFrame(X_test_array, columns=feature_names),
pd.DataFrame(shap_values[1], columns=[f"{x}_shap" for x in feature_names])
],
axis="columns"
).sort_index(axis="columns")
)
audit_dataframe.head(3)
| age | age_shap | cause_of_death_asphyxiated_restrained | cause_of_death_asphyxiated_restrained_shap | cause_of_death_beaten_bludgeoned_with_instrument | cause_of_death_beaten_bludgeoned_with_instrument_shap | cause_of_death_burned_smoke_inhalation | cause_of_death_burned_smoke_inhalation_shap | cause_of_death_chemical_agent_pepper_spray | cause_of_death_chemical_agent_pepper_spray_shap | ... | race_hispanic_latino | race_hispanic_latino_shap | race_middle_eastern | race_middle_eastern_shap | race_native_american_alaskan | race_native_american_alaskan_shap | race_race_unspecified | race_race_unspecified_shap | symptoms_of_mental_illness | symptoms_of_mental_illness_shap | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -0.534156 | -0.039886 | 0.0 | -0.007479 | 0.0 | -0.001080 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.003194 | 0.0 | 0.0 | 0.0 | -0.001401 | 1.0 | 0.030105 | 0.0 | -0.007136 |
| 1 | -0.820163 | -0.012115 | 0.0 | -0.008413 | 0.0 | -0.001327 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | -0.002723 | 0.0 | 0.0 | 0.0 | -0.001538 | 0.0 | -0.012061 | 1.0 | -0.011863 |
| 2 | -0.319651 | -0.037459 | 0.0 | -0.008016 | 0.0 | -0.001250 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.002469 | 0.0 | 0.0 | 0.0 | -0.001629 | 0.0 | -0.014678 | 0.0 | -0.009156 |
3 rows × 56 columns
Define two sample t-test that tests the relative impact of a variable on the output probability of the model
def hypothesis_accident_probability(feature, increases=True):
relationship = "greater_than" if increases else "less_than"
return {
feature: Column(checks=Check.isin([1, 0])),
f"{feature}_shap": Column(
pa.Float,
checks=Hypothesis.two_sample_ttest(
sample1=1,
sample2=0,
groupby=feature,
relationship=relationship,
alpha=0.01,
)
),
}
Programmatically construct the schema and validate feature_shap_df.
columns = {}
# increases probability of disposition "accidental"
for column in [
"cause_of_death_vehicle",
"cause_of_death_tasered",
"cause_of_death_asphyxiated_restrained",
"cause_of_death_medical_emergency",
"cause_of_death_drug_overdose",
"race_race_unspecified",
"race_native_american_alaskan",
]:
columns.update(hypothesis_accident_probability(column, increases=True))
# decreases probability of disposition "accidental"
for column in [
"cause_of_death_gunshot",
"race_european_american_white",
"race_asian_pacific_islander",
]:
columns.update(hypothesis_accident_probability(column, increases=False))
model_audit_schema = pa.DataFrameSchema(columns)
try:
model_audit_schema(audit_dataframe)
print("Model audit results pass! ✅")
except pa.errors.SchemaError as exc:
print("Model audit results fail ❌")
print(exc)
Model audit results pass! ✅
More Questions 🤔¶
- Why would
race_unspecifiedbe associated with a higher probability ofaccidentalrulings? - Can we predict/impute the
dispositionof unreported, unknown, or pending cases? What would that get us? - What's the generative process by which these data are being created and collected?
- Are the interaction effects between different demographic variables, e.g.
race_african_american_blackand other variables?
Takeaways¶
- Data validation is a means to multiple ends: reproducibility, readability, maintainability, and statistical type safety
- It's an iterative process between exploring the data, acquiring domain knowledge, and writing validation code.
panderaschemas are executable contracts that enforce the statistical properties of a dataframe at runtime and can be flexibly interleaved with data processing and analysis logic.
panderadoesn't automate data exploration or the data validation process. The user is responsible for identifying which parts of the pipeline are critical to test and defining the contracts under which data are considered valid.
Experimental Features¶
Schema Inference
schema = pa.infer_schema(dataframe)
Schema Serialization
pa.io.to_yaml(schema, "schema.yml")
pa.io.to_script(schema, "schema.py")
Roadmap: Feature Proposals¶
Define domain-specific schemas, types, and checks, e.g. for machine learning
# validate a regression model dataset
schema = pa.machine_learning.supervised.TabularSchema(
targets={"regression": pa.TargetColumn(type=pa.ml_dtypes.Continuous)},
features={
"continuous": pa.FeatureColumn(type=pa.ml_dtypes.Continuous),
"categorical": pa.FeatureColumn(type=pa.ml_dtypes.Categorical),
"ordinal": pa.FeatureColumn(type=pa.ml_dtypes.Ordinal),
}
)
Generate synthetic data based on schema definition as constraints
dataset = schema.generate_samples(100)
X, y = dataset[schema.features], dataset[schema.targets]
estimator.fit(X, y)
Contributions are Welcome!¶
Repo: https://github.com/pandera-dev/pandera
- Improving documentation
- Submit feature requests (e.g. additional built-in
CheckandHypothesismethods) - Submit bugs/issues or pull requests on Github
